Introduction
This tutorial shows the basics of how to use the package to calculate and visualize several metrics using map based inputs. It utilizes the package’s built-in data, which is the same example data used in Fletcher et al. (2019).
Load the Data
# "Load" the data. In this case we are using data built into the package.
# In practice, users will likely load raster data using the raster() function
# from the raster package.
res_data <- samc::example_split_corridor$res
abs_data <- samc::example_split_corridor$abs
init_data <- samc::example_split_corridor$init
# To make things easier for plotting later, convert the matrices to rasters
res_data <- samc::rasterize(res_data)
abs_data <- samc::rasterize(abs_data)
init_data <- samc::rasterize(init_data)
# Plot the data and make sure it looks good. The built-in data is in matrices,
# so we use the raster() function to help with the plotting. Note that when
# matrices are used by the package, it sets the extents based on the number of
# rows/cols. We do the same thing here when converting to a raster, otherwise
# the default extents will be (0,1) for both x and y, which is not only
# uninformative, but can result in "stretching" when visualizing datasets
# based non-square matrices.
plot(res_data, main = "Example Resistance Data", xlab = "x", ylab = "y", col = viridis(256))
plot(abs_data, main = "Example Absorption Data", xlab = "x", ylab = "y", col = viridis(256))
plot(init_data, main = "Example Occupancy Data", xlab = "x", ylab = "y", col = viridis(256))


Create the samc Object
# Setup the details for our transition function
rw_model <- list(fun = function(x) 1/mean(x), # Function for calculating transition probabilities
dir = 8, # Directions of the transitions. Either 4 or 8.
sym = TRUE) # Is the function symmetric?
# Create a `samc-class` object using the resistance and absorption data. We use the
# recipricol of the arithmetic mean for calculating the transition matrix. Note,
# the input data here are matrices, not RasterLayers.
samc_obj <- samc(res_data, abs_data, model = rw_model)
# Print out the samc object and make sure everything is filled out. Try to
# double check some of the values, such as the nrows/ncols of the landscape
# data. The dimensions of the matrix (slot p) should be the number of non-NA
# cells in your data +1. In this case, our data has 2624 non-NA cells, so the
# matrix should be 2625 x 2625
str(samc_obj)
#> Formal class 'samc' [package "samc"] with 15 slots
#> ..@ data :Formal class 'samc_data' [package "samc"] with 3 slots
#> .. .. ..@ f :Formal class 'dgCMatrix' [package "Matrix"] with 6 slots
#> .. .. .. .. ..@ i : int [1:21612] 0 1 115 116 117 0 1 2 116 117 ...
#> .. .. .. .. ..@ p : int [1:2625] 0 5 11 17 23 29 35 41 47 53 ...
#> .. .. .. .. ..@ Dim : int [1:2] 2624 2624
#> .. .. .. .. ..@ Dimnames:List of 2
#> .. .. .. .. .. ..$ : NULL
#> .. .. .. .. .. ..$ : NULL
#> .. .. .. .. ..@ x : num [1:21612] 1 -0.226 -0.138 -0.163 -0.104 ...
#> .. .. .. .. ..@ factors : list()
#> .. .. ..@ t_abs: num [1:2624] 2e-04 2e-04 2e-04 2e-04 2e-04 ...
#> .. .. ..@ c_abs: num[0 , 0 ]
#> ..@ conv_cache: NULL
#> ..@ model :List of 4
#> .. ..$ fun :function (x)
#> .. ..$ dir : num 8
#> .. ..$ sym : logi TRUE
#> .. ..$ name: chr "RW"
#> ..@ source : chr "SpatRaster"
#> ..@ nodes : int 2624
#> ..@ map :S4 class 'SpatRaster' [package "terra"]
#> ..@ crw_map : NULL
#> ..@ prob_mat : NULL
#> ..@ names : NULL
#> ..@ clumps : int 1
#> ..@ override : logi FALSE
#> ..@ solver : chr "direct"
#> ..@ threads : num 1
#> ..@ precision : chr "double"
#> ..@ .cache :<environment: 0x564274b1f068>Basic Analysis
# Convert the initial state data to probabilities
init_prob_data <- init_data / sum(values(init_data), na.rm = TRUE)
# Calculate short- and long-term mortality metrics and long-term dispersal
short_mort <- mortality(samc_obj, init_prob_data, time = 4800)
long_mort <- mortality(samc_obj, init_prob_data)
long_disp <- dispersal(samc_obj, init_prob_data)
#>
#> Cached diagonal not found.
#> Performing setup. This can take several minutes... Complete.
#> Calculating matrix inverse diagonal...
#> Computing: 100% (done)
#> Complete
#> Diagonal has been cached. Continuing with metric calculation...Visualization
# Create rasters using the vector result data for plotting.
short_mort_map <- map(samc_obj, short_mort)
long_mort_map <- map(samc_obj, long_mort)
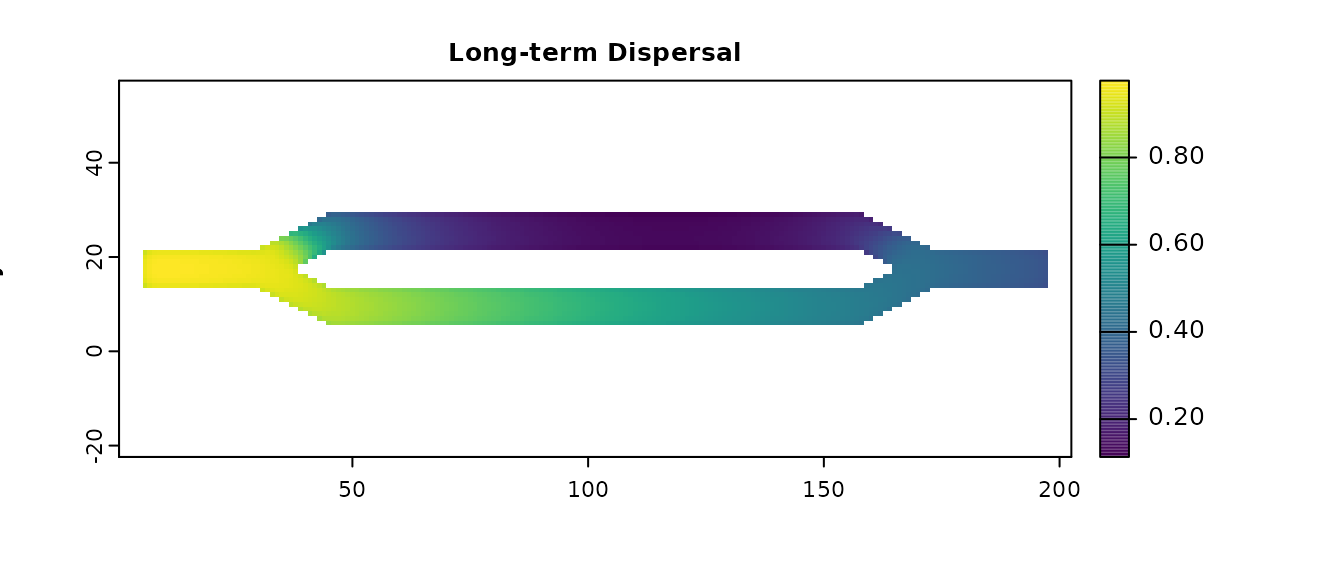
long_disp_map <- map(samc_obj, long_disp)
# Plot the mortality and dispersal results
plot(short_mort_map, main = "Short-term Mortality", xlab = "x", ylab = "y", col = viridis(256))
plot(long_mort_map, main = "Long-term Mortality", xlab = "x", ylab = "y", col = viridis(256))
plot(long_disp_map, main = "Long-term Dispersal", xlab = "x", ylab = "y", col = viridis(256))